vignettes/developers/bindings.Rmd
bindings.RmdWriting Bindings
When writing bindings between C++ compute functions and R functions, the aim is to expose the C++ functionality via the same interface as existing R functions. The syntax and functionality should match that of the existing R functions (though there are some exceptions) so that users are able to use existing tidyverse or base R syntax, whilst taking advantage of the speed and functionality of the underlying arrow package.
One of main ways in which users interact with arrow is via dplyr syntax called on Arrow objects. For example, when a user calls dplyr::mutate() on an Arrow Tabular, Dataset, or arrow data query object, the Arrow implementation of mutate() is used and under the hood, translates the dplyr code into Arrow C++ code.
When using dplyr::mutate() or dplyr::filter(), you may want to use functions from other packages. The example below uses stringr::str_detect().
## # A tibble: 2 x 14
## name height mass hair_c~1 skin_~2 eye_c~3 birth~4 sex gender homew~5
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
## 1 Darth Vader 202 136 none white yellow 41.9 male mascu~ Tatooi~
## 2 Darth Maul 175 80 none red yellow 54 male mascu~ Dathom~
## # ... with 4 more variables: species <chr>, films <list>, vehicles <list>,
## # starships <list>, and abbreviated variable names 1: hair_color,
## # 2: skin_color, 3: eye_color, 4: birth_year, 5: homeworld
## # i Use `colnames()` to see all variable namesThis functionality has also been implemented in Arrow, e.g.:
library(arrow)
arrow_table(starwars) %>%
filter(str_detect(name, "Darth")) %>%
collect()## # A tibble: 2 x 14
## name height mass hair_c~1 skin_~2 eye_c~3 birth~4 sex gender homew~5
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
## 1 Darth Vader 202 136 none white yellow 41.9 male mascu~ Tatooi~
## 2 Darth Maul 175 80 none red yellow 54 male mascu~ Dathom~
## # ... with 4 more variables: species <chr>, films <list<character>>,
## # vehicles <list<character>>, starships <list<character>>, and abbreviated
## # variable names 1: hair_color, 2: skin_color, 3: eye_color, 4: birth_year,
## # 5: homeworld
## # i Use `colnames()` to see all variable namesThis is possible as a binding has been created between the call to the stringr function str_detect() and the Arrow C++ code, here as a direct mapping to match_substring_regex. You can see this for yourself by inspecting the arrow data query object without retrieving the results via collect().
arrow_table(starwars) %>%
filter(str_detect(name, "Darth"))## Table (query)
## name: string
## height: int32
## mass: double
## hair_color: string
## skin_color: string
## eye_color: string
## birth_year: double
## sex: string
## gender: string
## homeworld: string
## species: string
## films: list<item: string>
## vehicles: list<item: string>
## starships: list<item: string>
##
## * Filter: match_substring_regex(name, {pattern="Darth", ignore_case=false})
## See $.data for the source Arrow objectIn the following sections, we’ll walk through how to create a binding between an R function and an Arrow C++ function.
Walkthrough
Imagine you are writing the bindings for the C++ function starts_with() and want to bind it to the (base) R function startsWith().
First, take a look at the docs for both of those functions.
Examining the R function
Here are the docs for R’s startsWith() (also available at https://stat.ethz.ch/R-manual/R-devel/library/base/html/startsWith.html)

It takes 2 parameters; x - the input, and prefix - the characters to check if x starts with.
Examining the C++ function
Now, go to the compute function documentation and look for the Arrow C++ library’s starts_with() function:

The docs show that starts_with() is a unary function, which means that it takes a single data input. The data input must be a string-like class, and the returned value is boolean, both of which match up to R’s startsWith().
There is an options class associated with starts_with() - called MatchSubstringOptions - so let’s take a look at that.
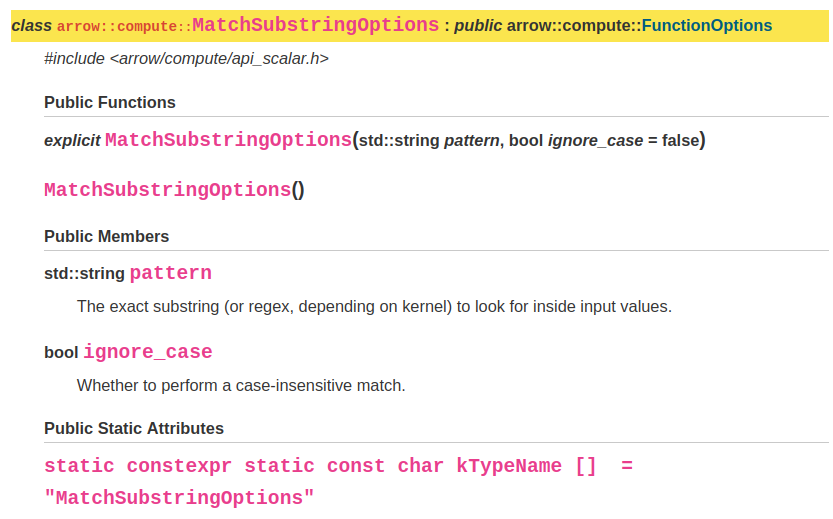
Options classes allow the user to control the behaviour of the function. In this case, there are two possible options which can be supplied - pattern and ignore_case, which are described in the docs shown above.
Comparing the R and C++ functions
What conclusions can be drawn from what you’ve seen so far?
Base R’s startsWith() and Arrow’s starts_with() operate on equivalent data types, return equivalent data types, and as there are no options implemented in R that Arrow doesn’t have, this should be fairly simple to map without a great deal of extra work.
As starts_with() has an options class associated with it, we’ll need to make sure that it’s linked up with this in the R code.
In case you’re wondering about the difference between arguments in R and options in Arrow, in R, arguments to functions can include the actual data to be analysed as well as options governing how the function works, whereas in the C++ compute functions, the arguments are the data to be analysed and the options are for specifying how exactly the function works.
So let’s get started.
Step 1 - add unit tests
We recommend a test-driven-development approach - write failing tests first, then check that they fail, and then write the code needed to make them pass. Thinking up-front about the behavior which needs testing can make it easier to reason about the code which needs writing later.
Look up the R function that you want to bind the compute kernel to, and write a set of unit tests that use a dplyr pipeline and compare_dplyr_binding() (and perhaps even compare_dplyr_error() if necessary. These functions compare the output of the original function with the dplyr bindings and make sure they match.
We recommend looking at the documentation next to the source code for these functions to get a better understanding of how they work.
You should make sure you’re testing all parameters of the R function in your tests.
Below is a possible example test for startsWith().
Step 2 - Hook up the compute function with options class if necessary
If the C++ compute function can have options specified, make sure that the function is linked with its options class in make_compute_options() in the file arrow/r/src/compute.cpp. You can find out if a compute function requires options by looking in the docs here: https://arrow.apache.org/docs/cpp/compute.html
In the case of starts_with(), it looks something like this:
if (func_name == "starts_with") {
using Options = arrow::compute::MatchSubstringOptions;
bool ignore_case = false;
if (!Rf_isNull(options["ignore_case"])) {
ignore_case = cpp11::as_cpp<bool>(options["ignore_case"]);
}
return std::make_shared<Options>(cpp11::as_cpp<std::string>(options["pattern"]),
ignore_case);
}You can usually copy and paste from a similar existing example. In this case, as the option ignore_case doesn’t map to any parameters of startsWith(), we give it a default value of false but if it’s been set, use the set value instead. As the pattern argument maps directly to prefix in startsWith() we can pass it straight through.
Step 3 - Map the R function to the C++ kernel
The next task is writing the code which binds the R function to the C++ kernel.
Step 3a - See if direct mapping is appropriate
Compare the C++ function and R function. If they are simple functions with no options, it might be possible to directly map between the C++ and R in unary_function_map, in the case of compute functions that operate on single columns of data, or binary_function_map for those which operate on 2 columns of data.
As startsWith() requires options, direct mapping is not appropriate.
Step 3b - If direct mapping not possible, try a modified implementation
If the function cannot be mapped directly, some extra work may be needed to ensure that calling the arrow version of the function results in the same result as calling the R version of the function. In this case, the function will need adding to the nse_funcs function registry. Here is how this might look for startsWith():
register_binding("base::startsWith", function(x, prefix) {
Expression$create(
"starts_with",
x,
options = list(pattern = prefix)
)
})In the source files, all the register_binding() calls are wrapped in functions that are called on package load. These are separated into files based on subject matter (e.g., R/dplyr-funcs-math.R, R/dplyr-funcs-string.R): find the closest analog to the function whose binding is being defined and define the new binding in a similar location. For example, the binding for startsWith() is registered in dplyr-funcs-string.R next to the binding for endsWith().
Note: we use the namespace-qualified name (i.e. "base::startsWith") for a binding. This will register the same binding both as startsWith() and as base::startsWith(), which will allow us to use the pkg:: prefix in a call.
arrow_table(starwars) %>%
filter(stringr::str_detect(name, "Darth"))## Table (query)
## name: string
## height: int32
## mass: double
## hair_color: string
## skin_color: string
## eye_color: string
## birth_year: double
## sex: string
## gender: string
## homeworld: string
## species: string
## films: list<item: string>
## vehicles: list<item: string>
## starships: list<item: string>
##
## * Filter: match_substring_regex(name, {pattern="Darth", ignore_case=false})
## See $.data for the source Arrow objectHint: you can use call_function() to call a compute function directly from R. This might be useful if you want to experiment with a compute function while you’re writing bindings for it, e.g.
call_function(
"starts_with",
Array$create(c("Apache", "Arrow", "R", "package")),
options = list(pattern = "A")
)## Array
## <bool>
## [
## true,
## true,
## false,
## false
## ]Step 4 - Run (and potentially add to) your tests.
In the process of implementing the function, you will need at least one test to make sure that your binding works and that future changes to the Arrow R package don’t break it! Bindings are tested in files that correspond to the file in which they were defined (e.g., startsWith() is tested in tests/testthat/test-dplyr-funcs-string.R) next to the tests for endsWith().
You may end up implementing more tests, for example if you discover unusual edge cases. This is fine - add them to the ones you wrote originally, and run them all. If they pass, you’re done and you can submit a PR. If you’ve modified the C++ code in the R package (for example, when hooking up a binding to its options class), you should make sure to run arrow/r/lint.sh to lint the code.